sgRNA设计
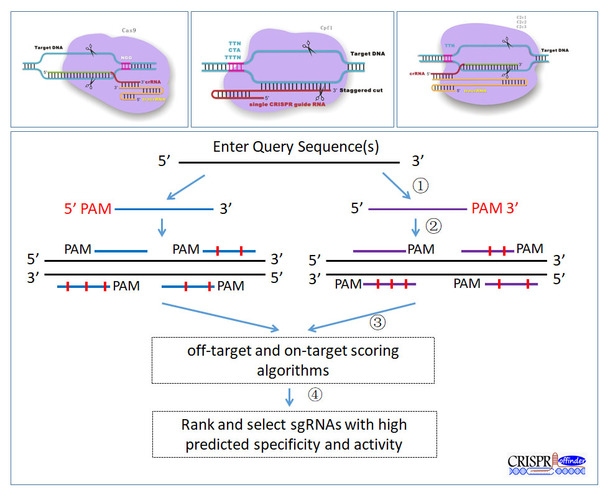
主要流程
1.全基因组搜索sgRNA
2.根据参考文件对sgRNA进行注释
2.1提取评分等级为一下的sgRNA ID信息
2.2合并所有的sgRNA信息
2.3去除脱靶的sgRNA

3.sgRNA筛选
4.参考
Last updated


Last updated
perl sgRNAcas9_3.0.5.pl
-i genes_end.fasta
-x 20
-l 40
-m 60
-g Ghirsutum_genome.fasta
-o b
-t s -v l -n 5 -s -3 -e 33-i <str> Input file
-x <int> Length of sgRNA[20]
-l <int> The minimum value of GC content [20]
-m <int> The maximum value of GC content [80]
-g <str> The reference genome sequence
-o <str> Searching CRISPR target sites using DNA strands based option(s/a/b)
[s, sense strand searching mode]
[a, anti-sense strand searching mode]
[b, both strand searching mode]
-t <str> Type of sgRNA searching mode(s/p)
[s, single-gRNA searching mode]
[p, paired-gRNA searching mode]
-v <str> Operation system(w/l/u/m/a)
[w, for windows-32, 64]
[l, for linux-64]
[u, for linux-32]
[m, for MacOSX-64]
[a, for MacOSX-32]
-n <int> Maximum number of mismatches [5]
-s <int> The minimum value of sgRNA offset [-2] 错配罚分
-e <int> The maximum value of sgRNA offset [32]
-p <str> Output path#M表示错配碱基数
0M 1M 2M 3M rank
2 0 0 0 repeat_sites_or_bad
1 0 0 5 low_risk
1 0 0 3 Bestperl ../Usefull_Script/ot2gtf_v2.pl -i Low_OT.text -g ../Gh_gene.gtf -o Low_OT_gtf_out.text
#也可以将上一步所有的OT文件合并之后,在运行脚本#awk脚本
-F "\t" '{
a=substr($1,1,15);a1=substr($1,6,1);a2=substr($1,7,2);b1=substr($5,6,1);b2=substr($5,7,2)
}{
if(a==$8)print $0"\t"0;
else if(a1=="A"&&b1=="D"){
if(a2==b2||a2=="02"&&b2==03||a2=="03"&&b2=="02"){
print $0"\t"0;}else print $0"\t"1;}
else if(a1=="D"&&b1=="A"){
if(a2==b2||a2=="02"&&b2=="03"||a2=="03"&&b2=="02"){
print $0"\t"0;}else print $0"\t"1;}
else print $0"\t"1;}' Best_Repeat_Low_OT_gtf >2222222usage:
-h|--help print help information
-g|--gff= gff file path way
-s|--sgRNA= sgRNA file path way
-l|--genelength= length of gene
-r|--sequence= sgRNA sequence path way
-o|--outfile= output file path way