表观数据分析
转座子数据
## 提取所有intron坐标
python ~/software/hisat2-2.1.0/extract_splice_sites.py ~/work/Alternative/result/Ga_result/CO11_12_result/06_Alignment/all.collapsed.gtf >1
awk '{print $1,$2+2,$3,$4}' OFS="\t" 1 >all_intron.bed
## 提取AS对应事件的Intron坐标
cp ../../Position/IR/A2_all.bed >IR.bed
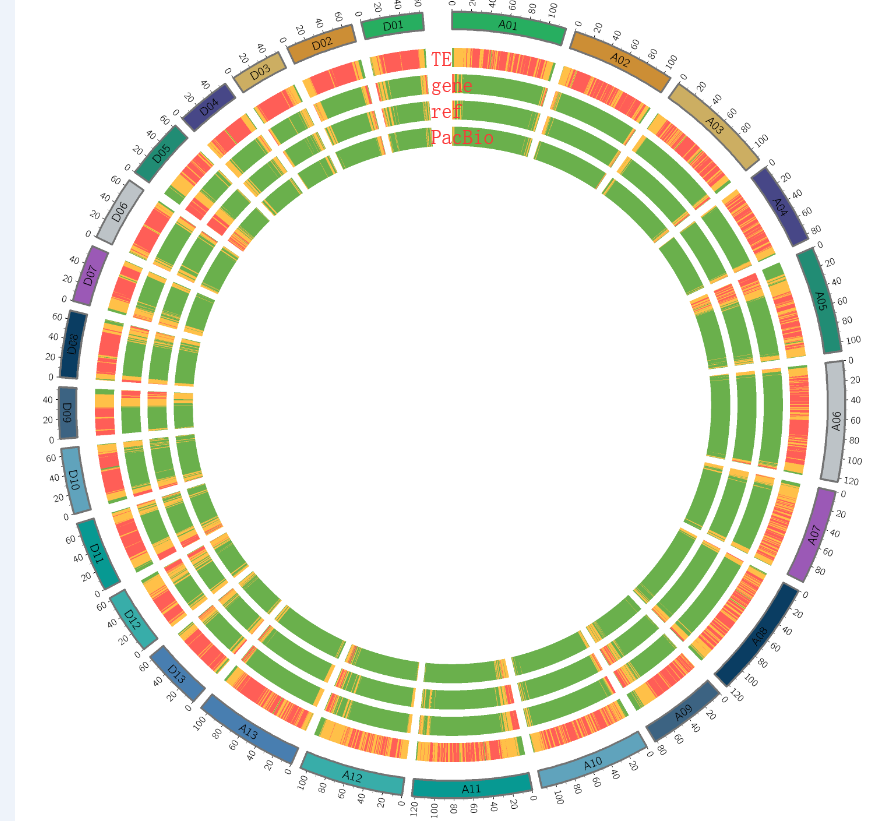
## 取交集Circos图数据
统计每个区域的覆盖度,而不是数目
制作染色体文件
甲基化数据
换另外一种计算方法
Last updated