pre程序
pre输入文件格式
<str1> <chr1> <pos1> <frag1> <str2> <chr2> <pos2> <frag2>python脚本
Pre转化格式
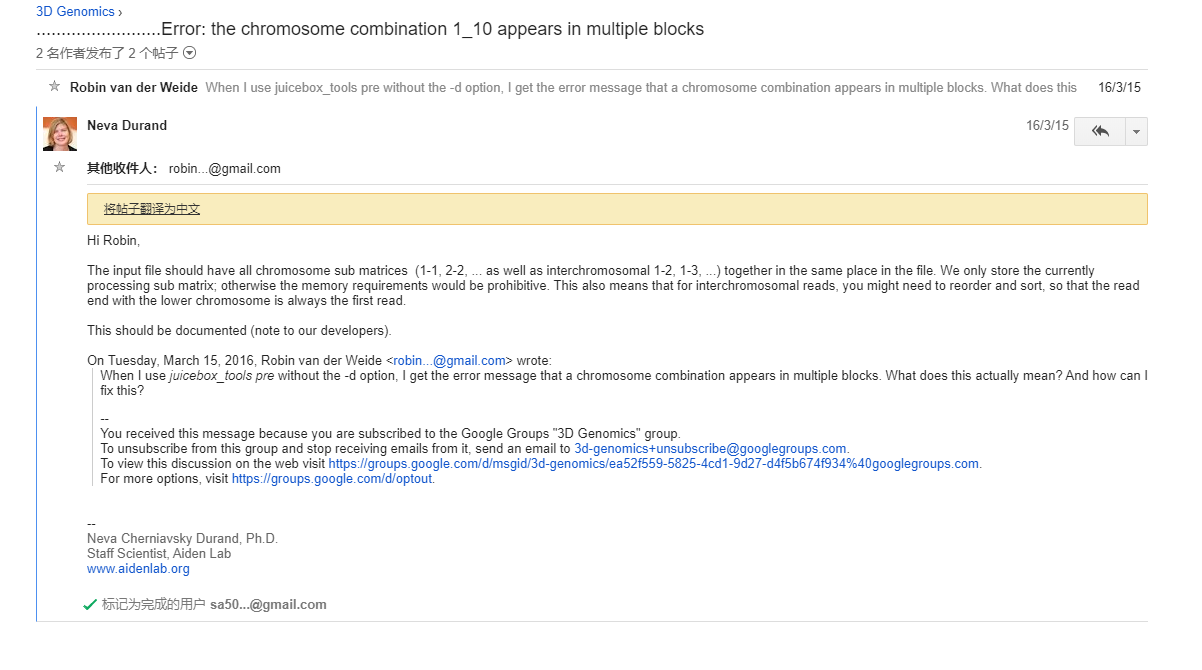
报错
测试数据
pre提交LSF任务
HiCCUPs任务
Last updated
<str1> <chr1> <pos1> <frag1> <str2> <chr2> <pos2> <frag2>
Last updated
python ~/github/zpliuCode/Hi-c/HiCProTojuicer.py -i ./rawdata.allValidPairs -s Scaffold -t 20 -o ~/github/juicer/LSF/test/Gb_HiC-Pro.txt
## 提交任务
bsub -q high -M 200G -R span[hosts=1] -J Hic-Pro -n 20 -e %J.err -o %J.out "python ~/github/zpliuCode/Hi-c/HiCProTojuicer.py -i /data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_0D_HiC_merged_results/hic_results/data/rawdata/rawdata.allValidPairs -s Scaffold -t 20 -o ~/github/juicer/LSF/test/Gb_HiC-Pro.txt"## 查看文件行数
wc -l 文件
## 分割文件
split -行数 文件名
## 批量提交脚本java -jar scripts/juicer_tools.jar pre -r 10000 data/Gb_HiC-Pro.txt data/Gb.hic Gb_HAU_v2.0_Chrosome.txt........Error: the chromosome combination 1_1 appears in multiple blocks#0D~25D的数据和叶片的数据
/data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_0D_HiC_merged_results/hic_results/data/rawdata
/data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_5D_HiC_merged_results/hic_results/data/rawdata
/data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_10D_HiC_merged_results/hic_results/data/rawdata
/data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_20D_HiC_merged_results/hic_results/data/rawdata
## 叶片数据
/data/cotton/MaojunWang/WMJ_fiber3Dgenome/Gb379_leaf_HiC_merged_results/hic_results/data/rawdatabsub -q high -J pre -e %J.err -o %J.out -n 10 -R span[hosts=1] "java -jar ~/github/juicer/LSF/scripts/juicer_tools.jar pre -r 10000 --threads 10 ./Gb379_5D_HiC_raw.txt ./Gb379_5D_HiC_raw.hic ~/github/juicer/LSF/Gb_HAU_v2.0_Chrosome.txt "for i in `ls ./|grep hic`
do
b=`echo $i|sed 's/raw\.hic//g'`loop
bsub -q gpu -J pre -e %J.err -o %J.out -n 5 -R span[hosts=1] "java -jar ~/github/juicer/LSF/scripts/juicer_tools.jar hiccups -m 500 -r 10000 -f 0.1 -p 2 -i 5 -d 20000 -k KR --threads 5 ./${i} ./${b}"
done
##使用默认参数跑一下
bsub -q gpu -J pre -e %J.err -o %J.out -n 5 -R span[hosts=1] "java -jar ~/github/juicer/LSF/scripts/juicer_tools.jar hiccups -m 500 -r 5000,10000 -d 20000,21000 -k KR -f .1,.1 -p 4,2 -i 7,5 -t 0.02,1.5,1.75,2 --threads 5 -c Gbar_A01,Gbar_A02,Gbar_A03,Gbar_A04,Gbar_A05,Gbar_A06,Gbar_A07,Gbar_A08,Gbar_A09,Gbar_A10,Gbar_A11,Gbar_A12,Gbar_A13,Gbar_D01,Gbar_D02,Gbar_D03,Gbar_D04,Gbar_D05,Gbar_D06,Gbar_D07,Gbar_D08,Gbar_D09,Gbar_D10,Gbar_D11,Gbar_D12,Gbar_D13 ./${i} ./${b}"
bsub -q gpu -J pre -e %J.err -o %J.out -n 10 -R span[hosts=1] "java -jar ~/github/juicer/LSF/scripts/juicer_tools.jar hiccups -m 500 -r 10000,25000 --threads 10 ./${i} ./${b}"